- Basic Veterinary Science
- Veterinary Anatomy and Histology
- Veterinary Pharmacology
- Veterinary Physiology and Biochemistry
- Biomedical Science-Developmental Genetics
- Veterinary Microbiology
- Pathogenetic and Preventive Veterinary Science
- Veterinary Epidemiology
- Veterinary Pathology
- Veterinary Hygiene
- Veterinary Public Health
- Veterinary Parasitology
- Clinical Veterinary Science
- Veterinary Surgery
- Veterinary Radiology
- Veterinary Internal Medicine
- Veterinary Clinical Pathology
- Veterinary Theriogenology
- Preventive Physiology and Management
- Large Animal Clinic
- Veterinary Diagnosis and Development Center
- International Research Center for One Welfare
- Animal Medical Center
- National BioResource Project Paramecium
- Yamaguchi University Joint Graduate School of Veterinary Medicine
Molecular Immunology and Infectious Disease
<Science study on infection>
Faculty
NISHIGAKI Kazuo, PhD, DVM
Professor, Molecular Immunology and Infectious Disease
MIYAKE Ariko, PhD
Associate Professor, Laboratory of Molecular Immunology and Infectious Disease
Recruitment for the PhD course
Our laboratory is currently recruiting Ph.D. students. Although our primary research fields are infectious diseases, virology, and public health, we have also investigated a broad range of disciplines, including bioinformatics, zoology, immunology, and genetics. Our research subjects range from livestock, pets, and poultry to wild animals and humans. We welcome applicants from around the world and accept international students from diverse backgrounds. Please contact Professor Kazuo Nishigaki for inquiries. Thank you
- Riboflavin transporter: Evidence of a role as entry receptor for chimpanzee endogenous retrovirus
- The Mysterious Relationship Between Domestic Cats and Endogenous Retroviruses
- Placenta-specific Soluble Protein EnvV-Fca from Domestic Cats
- FeLIX is a restriction factor against mammalian retrovirus infections
- Resurrection: Ancient Retrovirus Revival
- Conservation and Management of Tsushima Leopard Cats
- Convergent evolution of antiviral machinery derived from ERV truncated envelope genes in multiple species
“Riboflavin transporter: Evidence of a role as entry receptor for chimpanzee endogenous retrovirus”
Key Research Points
- The ancient chimpanzee retrovirus (CERV1: Chimpanzee Endogenous Retrovirus 1) has caused widespread infections in primates over the past 10 million years.
- CERV1 became endogenous in great apes (chimpanzees, bonobos, and gorillas) and Old World monkeys.
- The receptor for CERV1 infection is a riboflavin transporter.
- CERV1 and porcine endogenous retroviruses (PERVs) belong to the same viral interference group and provide important insights into human xenotransplantation.
Endogenous retroviruses (ERVs) are remnants of ancient viral infections that integrated into the host genome and have passed across generations. These sequences serve as genetic fossils that offer insights into past viral infections and host-virus evolution. CERV1 is an extinct gammaretrovirus that was active in primates approximately 10 million years ago. Unlike other ERVs, ERVs are absent from humans and orangutans. This study aimed to determine how CERV1 infects host cells and becomes endogenized. The researchers identified that CERV1 utilizes riboflavin transporters, specifically SLC52A2 and SLC52A1, as entry receptors, a discovery that links ancient viral infection mechanisms to present-day cellular proteins.
Receptor screening using a human cDNA library identified the riboflavin transporter SLC52A2 as a key receptor for CERV1. Cells engineered to express either SLC52A2 or its homolog, SLC52A1 (from humans or chimpanzees), are susceptible to infection with CERV1-pseudotyped viruses, indicating that these transporters enable viral entry. Moreover, CERV1 shares these receptors with the porcine endogenous retrovirus (PERV), and viral interference assays have confirmed that CERV1 and PERV-A/C belong to the same interference group. This means that infection by one can block infection by the other, revealing an important aspect of host-virus interactions and competition for cellular entry.
CERV1 has been shown to infect a range of human and primate cell lines, particularly those with high SLC52A2 expression. In contrast, cells with low or no SLC52A1 expression remain permissive due to the abundance of SLC52A2. Phylogenetic and RNA-seq analyses revealed the widespread but varied expression of these transporters across tissues and species. Notably, CERV1 env genes have been found in the genomes of great apes and Old World monkeys, but not in humans or orangutans. This suggests that CERV1 circulates in primates after the divergence of these lineages. CERV1 env expression was detected in multiple tissues, with the highest levels in the lungs, thymus, and brain of different primates, implying co-evolution after endogenization.
This study provides crucial insights into how extinct retroviruses such as CERV1 infected their hosts, revealing riboflavin transporters as entry receptors. This highlights a case of convergent evolution in receptor usage by retroviruses and opens the door for a better understanding of host-virus co-evolution. Importantly, this study has implications for xenotransplantation, particularly for PERV, because the same receptors are involved. The lack of CERV1 in humans, despite its receptor expression, suggested that other resistance mechanisms, such as innate immune factors, might play a role. Overall, these findings deepened our understanding of ancient viral integration and contributed to fields such as palaeovirology, immunology, and evolutionary biology.
Information of published paper
Title: Riboflavin transporter: Evidence of a role as entry receptor for chimpanzee endogenous retrovirus
Authors: Loai AbuEeda#, Ariko Miyake#, Nashon Wanjala, Didik Pramono, Dimas Abdillah, Masanori Imamura, Masayuki Shimojima, Joachim Denner, Junna Kawasaki, Kazuo Nishigaki # These authors contributed equally to this work.
Journal: Virus Evolution
2025 May 7;11(1): veaf031.
DOI: https://doi.org/10.1093/ve/veaf031
“The Mysterious Relationship Between Domestic Cats and Endogenous Retroviruses”

Endogenous retroviruses (ERVs) are clusters of genetic sequences originating from ancient retroviral infections. Recently, our group discovered that an ERV known as ERV-DC8 in domestic cats retained both its infectivity and the ability to replicate autonomously, even after tens of thousands of years. Furthermore, when we examined domestic cats from various countries worldwide, we found that a high proportion carried ERV-DC8. This suggests that domestic cats and ERVs have a symbiotic relationship. To date, no ERVs with such infectivity or replication capacity have been found in humans.
The origin of domestic cats is believed to be traced back to the Middle East, where people began leading sedentary lifestyles and domesticating wild cats. It is speculated that people recognize the role of wildcats as predators that help keep grain storage areas free of pests. In addition, for a wildcat to become a “domesticated,” it would have been necessary to evolve a more docile nature suitable for living alongside humans. This behavioral adaptation likely played a key role in domestication. Domestic cats were formally classified as Felis silvestris catus, belonging to the order Carnivora, family Felidae, and genus Felis. They are affectionately known simply as “cats.” However, the origin of domestic cats remains unclear and their genome data differ across regions.
The genome contains a vast amount of genetic information, but only approximately 2% of the genome encodes proteins. Surprisingly, approximately 8% of the genomes of both cats and humans are composed of ERVs, which are immense genetic sequences originating from ancient retroviral infections. Retroviruses that currently circulate in humans and cats typically infect somatic cells. However, they become ERVs, which are genetic elements inherited from parent to offspring, if they infect germline cells and are passed on to the next generation.
Some ERVs are believed to have emerged over 10 million years ago, and have accumulated mutations, insertions, and deletions over time, rendering them essentially “scrap” or "junk DNA." In most cases, the ability of these sequences to form virus particles or remain infectious is lost. Thus, ERVs are considered to be molecular fossils of ancient infections. Studying them helps reveal the nature of viruses that once plagued ancient organisms and provides insight into virus-host coevolution, viral extinction mechanisms, and the evolutionary role of viruses in animals.
In the present study, we identified an infectious and self-replicating ERV-DC8 gene located on chromosome B1 in domestic cats. When this gene was introduced into cultured cells, ancient viral particles were produced. Since the ERV-DC8 gene sequence was intact, it retained the ability to form viral particles and remained infectious. Although the exact timing of ERV-DC integration into the cat genome remains unclear, it may date back as far as 2.7 million years. However, as the gene remains undamaged, it is estimated to have been integrated only over the past several tens of thousands of years.
When we examined the prevalence of ERV-DC8 in domestic cats from Africa, Europe, Asia, and Japan, it was found in 53–86% of cats. This integration likely occurred around the time wildcats evolved into domestic cats, and as human civilization progressed, domestic cats and ERV-DC8 spread globally. Such infectious and autonomously replicating ERVs have not been found in humans.
These findings suggested that while domestic cats carry a potentially dangerous virus at the genetic level, they manage and peacefully coexist with this ancient virus through sophisticated biological control mechanisms.
Information of published paper
Title: Endogenous retrovirus ERV-DC8 highly integrated in domestic cat populations is a replication-competent provirus
Authors: Didik Pramono#, Yutaro Muto#, Yo Shimazu, R.M.C. Deshapriya, Isaac Makundi, MariaCruz Arnal, Daniel Fernandez de Luco, Minh Ha Ngo, Ariko Miyake, Kazuo Nishigaki (#These authors contributed equally to this work)
Journal: Biochemical and Biophysical Research Communications
2024 Dec 17:738:150521.
DOI: 10.1016/j.bbrc.2024.150521
“Placenta-specific Soluble Protein EnvV-Fca from Domestic Cats ”
Summary
Endogenous retroviruses (ERVs) are remnants of ancestral viruses in host genomes. One feline ERV, EnV-Fca, was detected as a placenta-specific protein secreted by cells. Genetic analyses have shown that EnvV2 is widespread in vertebrates, with birds, bats, and rodents potentially involved in viral transmission. These findings provide a model of retroviral transmission and may elucidate the evolution of ERVs.
Endogenous retroviruses (ERVs) are ancient retroviral sequences that infect germ cells and are vertically transmitted to offspring of various vertebrate hosts, including mammals. The proviral genome is comprised of three major polyproteins: gag, pol, and env. Defective env-ERV refers to an Env protein that has a signal peptide and SU subunit, but contains a premature stop codon and lacks or has a partial deletion of the TM subunit. Defective env-ERV may be crucial for physiological functions in the host, such as maintaining homeostasis and placentation and acting as a restricting factor for infection. Other ERVs have been identified and are expressed in mammals as placenta-specific proteins. Syncytin is a captive retroviral envelope protein involved in the human placental function. Another placenta-specific protein, the group V member ERV Env (EnvV), has been identified. EnvVs can be classified into two groups, EnvV1 and EnvV2, which share a high degree of similarity.
We investigated the expression of ERV env genes in domestic cats in this study. EnvV in Felis catus (EnvV-Fca), is closely related to the EnvV2-Hum gene. EnvV-Fca is secreted by cells as a soluble protein. Additionally, EnV-Fca was specifically detected in placental trophoblast syncytiotrophobic layers. EnvV-Fca does not exhibit fusion activity, suggesting that it is not directly involved in syncytiotrophoblast formation during pregnancy or may be a restrictive factor for exogenous retroviral infection. Our findings revealed an intriguing evolutionary pattern of ERVs, showing that EnvV2 is not limited to mammals but extends to several non-mammalian vertebrates, such as birds, reptiles, fishes, and amphibians. However, we were unable to confirm the integration of EnvV2 into non-mammalian vertebrates in this study due to the limited information on non-mammalian vertebrates.
This study also built on bioinformatics data and TimeTree analysis. It was estimated that the integration time for the newly identified EnvV2 occurred at least 60 million years ago during mammalian evolution, particularly in rodents, elephants, and bats. Notably, as reported previously, bats, rodents, and birds are also involved in the transmission of retroviruses directly derived from mammalian retroviruses. This indicates the potential for the transmission of EnvV2 through these intermediate vectors. Moreover, as EnvV2 has a conserved immunosuppressive domain (ISD), it may have implications in immunotolerance during pregnancy.
Our findings presented a scenario of retroviral transmission among species that may have significant implications for elucidating retroviral transmission and ERV evolution. Furthermore, the present study provided a comprehensive characterization of EnvV-Fca, detailing the identification of the EnvV gene, investigation of its expression and possible function, and an evolutionary analysis of EnvV2 in mammalian and non-mammalian vertebrate hosts.
Information on published paper
Title: Characterization of the endogenous retrovirus-derived placenta-specific soluble protein EnvV-Fca from domestic cats
Authors: Didik Pramono, Kenji Sugimoto, Tohru Kimura, Ariko Miyake, and Kazuo Nishigaki.
Journal:
FEBS Letters
2024 Jul;598(14):1792-1806.
DOI: 10.1002/1873-3468.14873
“FeLIX is a restriction factor against mammalian retrovirus infections”
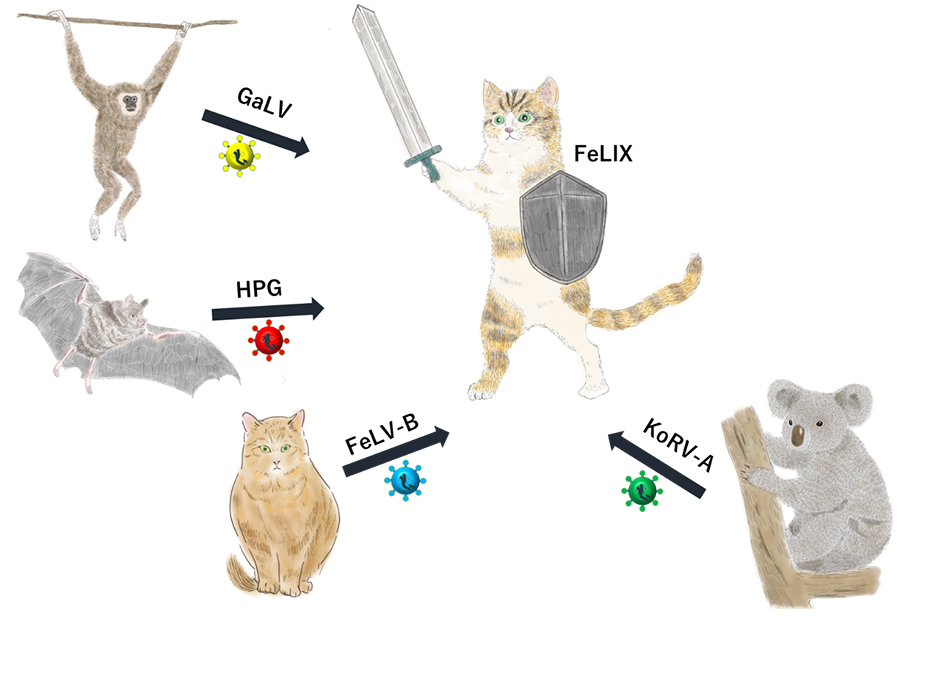
We identified a secreted protein that blocks various retroviral infections, including those caused by feline leukemia virus (FeLV), in domestic cats. The results of this study will facilitate the development of new methods for preventing and treating infectious diseases. FeLV is a retrovirus that causes anemia and hematopoietic diseases such as lymphoma and leukemia in domestic cats (Felis catus). This virus infects animals by binding to entry receptors on the cell surface. Notably, different FeLV subgroups have different entry receptors. For example, the entry receptor for FeLV-A is THTR1, a vitamin B1 transporter, whereas that of FeLV-B is Pit, a phosphate transporter.
FeLV-A is transmitted to domestic cats. Interestingly, FeLV-A infection led to viral gene recombination in domestic cats, resulting in the emergence of a new virus, FeLV-B. Unlike FeLV-A, FeLV-B is not transmitted to domestic cats. In this study, we investigated the mechanisms responsible for differences in the transmission of different viruses. Genome analysis revealed that the secreted protein FeLIX blocked the transmission of FeLV-B in domestic cats.
FeLIX is encoded by an endogenous retrovirus, known as the ancient virus, and is derived from its envelope gene. Genetic analysis revealed that FeLIX was derived from an ancient virus that infected domestic cat ancestors approximately 700,000 years ago. In addition to domestic cats, FeLIX has been detected in wild European cats (F. silvestris). Our findings revealed that FeLIX prevents FeLV-B infection by masking its entry receptors. FeLIX is a protein secreted by lymphocytes that circulates in the blood of domestic cats and inhibits the cellular entry of viruses. Additionally, we have observed retroviruses similar to FeLV-B in several animal species, including gibbons, bats, and koalas. Notably, FeLIX inhibited infections caused by these viruses. These findings suggested that FeLIX protects domestic cats against various retroviruses by acting as a restrictive agent.
Information of published paper
Title: FeLIX is a restriction factor against mammalian retrovirus infections Authors: Didik Pramono, Dai Takeuchi, Masato Katsuki, Loai AbuEed, Dimas Abdillah, Tohru Kimura, Junna Kawasaki, Ariko Miyake, and Kazuo Nishigaki Journal:
Journal of Virology
2024 Apr 16;98(4): e0177123.
DOI: https://doi.org/10.1128/jvi.01771-23
“Resurrection: Ancient Retrovirus Revival”
We discovered that infection with modern retroviruses in domestic cats could revive ancient retroviruses, resulting in the emergence of multiple new pathogenic viruses.
The genetic information (genomes) of humans and animals contain remnants of ancient retroviruses that infected their ancestors. These viruses are endogenous retroviruses (ERVs). ERVs make up approximately 8% of the human genome, and various types of ancient retroviral gene sequences, such as fossils, exist in the genome of domestic cats.
Ancient retroviruses, like their modern counterparts, are believed to have caused diseases in the ancestors of humans and animals. These infections occurred tens of thousands to millions of years ago. Over this vast timescale, the pathogenicity (disease-causing ability) of most ancient retroviruses was suppressed. However, some ERVs are still recognized to have pathogenic potential.
In this study, researchers discovered that feline leukemia virus (FeLV), a retrovirus that infects domestic cats, can recombine with ancient ERVs after infection, resulting in the emergence of new viruses that contribute to the development of lymphoma and leukemia. This study also showed that these recombinant viruses were derived from multiple types of ancient retroviruses.
These findings suggested that the genetic remnants of ancient viruses embedded in the genome for tens of thousands to millions of years may reawaken in the modern era through infection by contemporary viruses, posing a renewed threat.
Information of published paper
Title: Multiple Recombination Events between Endogenous Retroviral Elements and Feline Leukemia Virus
Authors: Minh Ha Ngo*, Loai AbuEed*, Junna Kawasaki*, Naoki Oishi, Didik Pramono*, Toru Kimura, Yu Sakurai, Kenji Watanabe, Yoichi Mizukami, Haruhi Ochi, Yukari Anai, Yuka Odawara, Daigo Umehara, Maki Kawamura, Shinya Watanabe, Ariko Miyake*, Kazuo Nishigaki
(*These authors contributed equally to this work)
Journal:
Journal of Virology
2024 Feb 20;98(2):e0140023.
DOI: 10.1128/jvi.01400-23
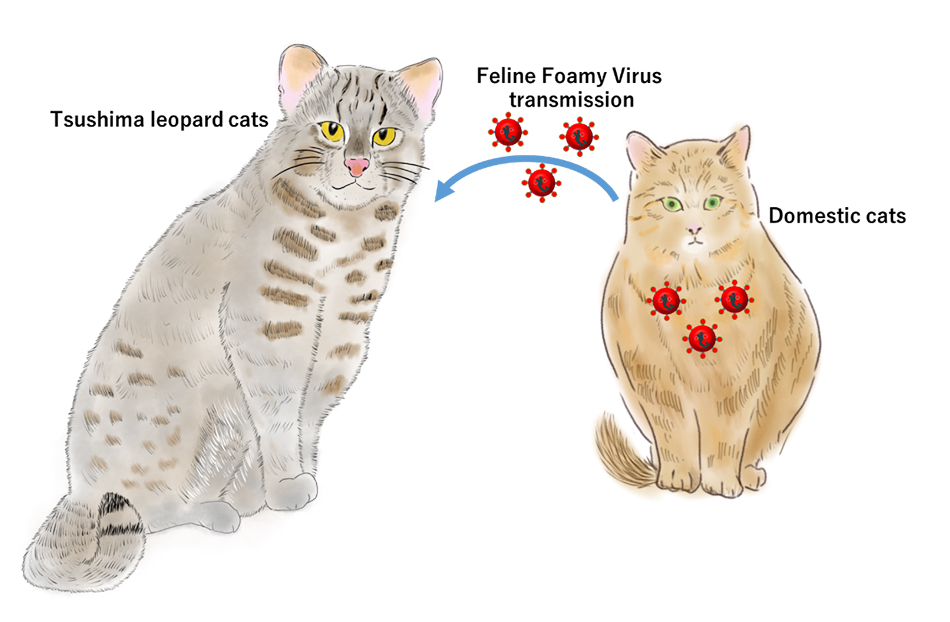
“Conservation and Management of Tsushima Leopard Cats”
Key Research Points
- Among 89 tested Tsushima leopard cats (Prionailurus bengalensis euptilurus), 7 individuals (7.86%) were infected with feline foamy virus (FFV), a type of retrovirus.
- Genetic sequencing and analysis revealed that the FFV strains infecting the Tsushima leopard cats matched those found in domestic cats (Felis catus) on Tsushima Island. This indicates that the virus originated in domestic cats and was transmitted to wild Tsushima leopard cats.
- Measures must be taken to prevent the transmission of infectious diseases from domestic cats to Tsushima leopard cats.
The Tsushima leopard cat (TLC, Prionailurus bengalensis euptilurus) is a critically endangered wild feline endemic to Tsushima Island in Japan. With a declining population of less than 100 individuals, this species faces multiple threats, including infectious diseases transmitted from domestic cats. One such concern is the feline foamy virus (FFV), a retrovirus commonly found in domestic cats (Felis catus) around the world. This study aimed to investigate whether FFV is present in TLCs and evaluate the potential for cross-species transmission from domestic cats. By analyzing the FFV prevalence, genotypes, and viral sequence similarities, this study provides key insights into the molecular epidemiology and conservation implications for endangered TLC.
A total of 89 TLC samples and 199 domestic cat samples from Tsushima Island were tested for FFV using nested PCR targeting the env gene (SU region). FFV was detected in 7 TLC (7.86%) and 28 domestic cat (14.07%) samples. FFV infection in domestic cats was significantly associated with co-infection with feline immunodeficiency virus (FIV) and feline gammaherpes virus 1 (FcaGHV1), but not with feline leukemia virus (FeLV). In particular, male cats were more likely to be infected with FIV and FcaGHV1, supporting the idea that viral transmission may occur through social behaviors such as biting and grooming. In contrast, there was no statistically significant association between FFV prevalence and sex or geographical location (Kamijima vs. Shimojima) in the TLCs.
Sequence analysis revealed that the FFV strains in the TLCs were genetically indistinguishable from those in domestic cats, particularly within the FUV-type clade. Phylogenetic trees constructed using SU region sequences showed that FFV isolates from domestic cats were spread across multiple clades, whereas all TLC isolates clustered within a single clade, indicating a likely spillover from domestic cats to TLCs. Two distinct FFV genotypes, F17/951-type and FUV-type, were identified in domestic cats, but only FUV-type was found in TLCs. Nucleotide similarities within genotypes were high (98–100%), whereas inter-genotype similarity ranged from 82–85%. The presence of both genotypes in domestic cats and only one genotype in the TLCs suggests that cross-species transmission was selective and recent.
This study is the first to report molecular evidence of FFV infection in Tsushima leopards and provides compelling evidence for interspecies viral transmission from domestic cats, likely through ecological overlap and behavioral contact. Given the endangered status of TLC and its limited geographic range, the introduction of retroviruses, such as FFV, could pose additional risks to population viability. This study highlights the urgent need for conservation strategies that reduce the contact between domestic and wild felines, such as managing stray cat populations, promoting indoor pet ownership, and incorporating FFV testing into routine wildlife disease surveillance. Finally, this study emphasizes the role of molecular epidemiology in wildlife conservation and the importance of the One Health approach in protecting biodiversity.
Information of published paper
Title: Feline Foamy Virus Transmission in Tsushima Leopard Cats (Prionailurus bengalensis euptilurus) on Tsushima Island, Japan
Authors: Loai AbuEed, Isaac Makundi, Ariko Miyake, Junna Kawasaki, Chisa Minoura, Yushi Koshida, Kazuo Nishigaki
Journal: Viruses, 2023 Mar 24;15(4):835.
DOI: 10.3390/v15040835
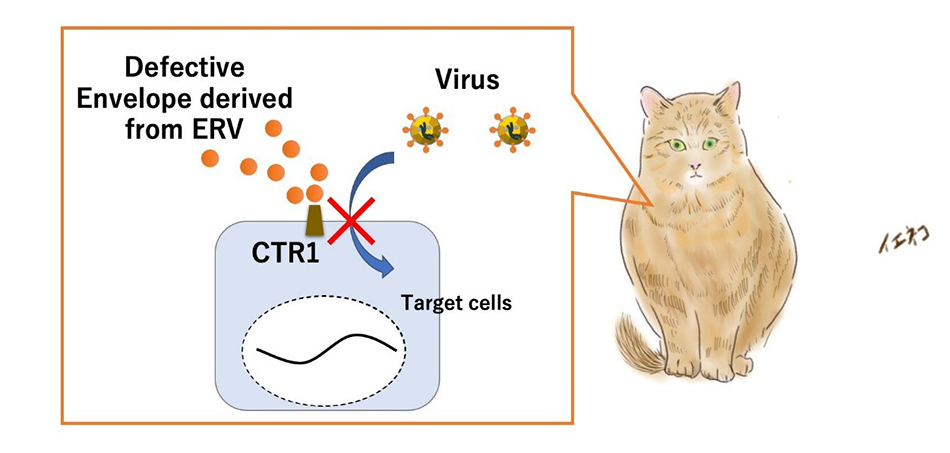
“Convergent evolution of antiviral machinery derived from ERV truncated envelope genes in multiple species”
Key Research Points
- The team elucidated the mechanism by which secreted proteins derived from the truncated envelope genes of ERVs inhibit viral infections by blocking the virus receptor CTR1 (copper transporter).
- They found that this ERV-derived viral defense mechanism exists in both feline and primate species, suggesting that it emerged independently in multiple animal species through convergent evolution.
- Because ERVs, which are remnants of ancient retroviral infections, are scattered throughout the genomes of both humans and animals, similar infection defense systems are likely universally present.
Human and animal genomes contain remnants of ancient horizontally transmitted retroviruses called endogenous retroviruses (ERVs), which constitute approximately 8% of the human genome. Although viruses do not fossilize like dinosaurs, the genetic remnants of ancient retroviruses that once infected our ancestors are preserved in our DNA, essentially as molecular fossils.
ERVs are found in both humans and animals. Similar to modern viruses, ancient viruses are also likely to cause diseases in their hosts. These infections occurred tens of thousands to millions of years ago. By studying these retroviruses, we can understand how humans and animals overcome viral threats over large timescales.
This study revealed that ancient retroviruses co-evolved with their hosts after infecting them. The resulting ERVs developed the ability to inhibit viral infections. In particular, the protein Refrex-1, which is found in domestic cats, is derived from the envelope gene of an ERV called ERV-DC. It is believed that ERV-DC infected the ancestors of domestic cats approximately 2.8 million years ago, and through co-evolution, Refrex-1 emerged.
Refrex-1 has evolved into a truncated secreted protein, meaning that it is released outside the cell. Although derived from a viral envelope, mutations shorten it and give it a new function; it binds to viral receptors on the cell surface and physically blocks viruses from entering the cells (Figure 2).
Convergent Evolution of Antiviral Systems
Refrex-1 acts as a physical barrier that interferes with viral entry and effectively prevents infection. The same type of antiviral defense system has been discovered in chimpanzees, bonobos, gorillas, macaques, and several primate species. These systems likely emerged as evolutionary responses to ancient viral infections. This study suggests that similar antiviral molecules appeared through convergent evolution across different species.
By retaining the DNA-level memory of ancient viral infections, animals not only defend themselves against new viral threats but may also contribute to the extinction of ancient viruses. Having similar antiviral systems across species may also enhance the resistance to viruses that can jump between species.
Information of published paper
Title: Convergent evolution of antiviral machinery derived from endogenous retrovirus truncated envelope genes in multiple species
Authors: Ariko Miyake*, Minh Ha Ngo*, Shelly Wulandari, Masayuki Shimojima, So Nakagawa, Junna Kawasaki, Kazuo Nishigaki
(*These authors contributed equally to this work)
Journal: Proceedings of the National Academy of Sciences of the United States of America (PNAS)
2022 Jun 28;119(26):e2114441119.
DOI: https://doi.org/10.1073/pnas.2114441119